The Legionella pneumophila lag-1 gene provides resistance to immune cells
New article on the discovery and functional analysis of a legionella pneumophila gene associated with isolates from human infections.
Our recent Nature Communications publication “Population analysis of Legionella pneumophila reveals a basis for resistance to complement-mediated killing” details the discovery and functional analysis of a legionella pneumophila gene associated with isolates from human infections. The analysis was acheived by a combination of computational and immunological methods and involved a close, long-term collaboration between Bryan Wee and Joana Alves.
Read the article abstract here:
"Legionella pneumophila is the most common cause of the severe respiratory infection known as Legionnaires’ disease. However, the microorganism is typically a symbiont of free-living amoeba, and our understanding of the bacterial factors that determine human pathogenicity is limited. Here we carried out a population genomic study of 902 L. pneumophila isolates from human clinical and environmental samples to examine their genetic diversity, global distribution and the basis for human pathogenicity. We find that the capacity for human disease is representative of the breadth of species diversity although some clones are more commonly associated with clinical infections. We identified a single gene (lag-1) to be most strongly associated with clinical isolates. lag-1, which encodes an O-acetyltransferase for lipopolysaccharide modification, has been distributed horizontally across all major phylogenetic clades of L. pneumophila by frequent recent recombination events. The gene confers resistance to complement-mediated killing in human serum by inhibiting deposition of classical pathway molecules on the bacterial surface. Furthermore, acquisition of lag-1 inhibits complement-dependent phagocytosis by human neutrophils, and promoted survival in a mouse model of pulmonary legionellosis. Thus, our results reveal L. pneumophila genetic traits linked to disease and provide a molecular basis for resistance to complement-mediated killing."
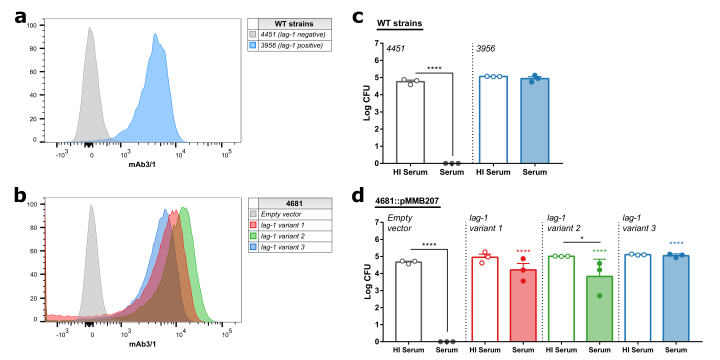
For more information, you can find the paper in this link: https://www.nature.com/articles/s41467-021-27478-z
Full Citation:
Wee BA, Alves J, Lindsay DSJ, Klatt AB, Sargison FA, Cameron RL, Pickering A, Gorzynski J, Corander J, Marttinen P, Opitz B, Smith AJ, Fitzgerald JR. Population analysis of Legionella pneumophila reveals a basis for resistance to complement-mediated killing. 10.1038/s41467-021-27478-z. PMID: 34887398.

