Computational approaches to study lncRNAs in vascular biology and disease
Considering the key role of long non-coding RNAs in gene expression, we aim to identify lncRNAs showing a change in expression during vasculature development or in pathology. The lncRNA regulation and potential mechanism of action is then investigated based on genome-wide data generated by our group or publicly available.
Genetic information is stored in the DNA of every cell in the form of gene unit. Some of the genes known as protein-coding genes contain information to code proteins and this happens through the production of a precursor RNA molecule. Our genome also contains non-coding genes that do not produce proteins but functional RNA molecules. Non-coding RNAs, in particular long non-coding RNAs, have gained considerable interests with their implication in development and disease in general as well as in the vasculature. These RNAs are also promising candidates for therapeutic approaches.
In this project, we are studying the RNA molecules that are present in different model of development of the vasculature or in samples from patients affected by cardiovascular diseases. The experiments obtained by our group or other scientist in the world generate complex data that requires extensive analysis. Our aim is to identify novel long non-coding RNAs that plays a role in vascular development and disease and predict their mode of action at the cellular level.
LncRNAs have received increasing attention in recent years and are now seen as key players in the regulation of gene expression. Their high specificity of expression also make them an interesting target for therapeutic approaches.
Identification of lncRNAs
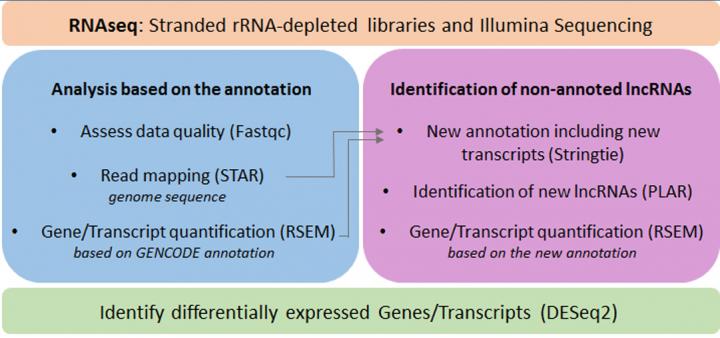
To identify lncRNAs differentially expressed and therefore with potential function during vascular biology or disease, we are using genome-wide transcriptome data (RNA-seq) of different biological models, for example:
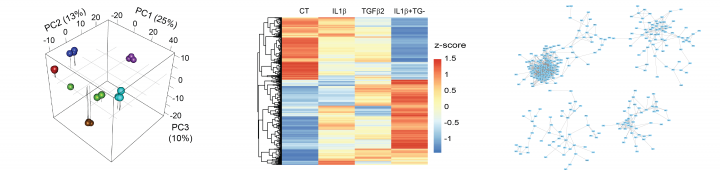
- We are performing RNAseq analysis was performed on cells undergoing endothelial to mesenchymal transition in vitro, a process observed in normal development as well as in pathology such as pulmonary hypertension.
Researchers: Axelle Caudrillier, Joe Monteiro, Julie Rodor, Lin Deng, Fatma Kok, Andrew Baker
- We are also analysing lncRNA expression in samples from patients with atherosclerosis. The unstable plaque blocking the artery wall is removed from the patients and we are comparing the “unstable” site of rupture from the “stable” adjacent region.
Researchers: John Hung, Julie Rodor, Margaret Ballantyne, Andrew Baker, David Newby, Jakub Kaczynsku
Because of their high specificity of expression, the catalogue of lncRNA genes in the genome is still incomplete. Our analysis is based on a pipeline called PLAR developed by our collaborator in Israel, Dr Igor Ultsky. It includes the quantification of annotated lncRNAs but also allows the discovery of novel lncRNAs.
Characterization of lncRNAs regulation and function
Despite the identification of numerous lncRNAs, the role of most lncRNAs is still unclear as their sequence does not allow to predict their function. LncRNAs can play very diverse roles by interacting with protein, DNA or RNA but the study of their mechanism of action is very challenging experimentally. In the nucleus, it has been reported that lncRNAs can regulate the transcription of other genes or affect RNA processing. In the cytoplasm, lncRNAs have been shown to control the stability of mRNAs, regulate transcription or act as decoy for miRNAs.
To predict lncRNA functions and guide downstream experiments, we are using different strategies based on high-throughput sequencing data (generated by our group or publicly available) as well as prediction tools.
Although lncRNAs are generally poorly conserved, some lncRNAs can be identified in several species and we hypothesize that lncRNA conservation would imply important molecular function. For this reason, we are developing tools to assess the conservation of lncRNAs, focusing on established animal model of vascular biology (such as pig, mouse and zebrafish) that could be used in the future to study the lncRNA function in vivo. We are working on this aspect in collaboration with the group of Professor Alan Archibald at the Roslin Institute.
Prediction of function for lncRNA often rely on a ‘guilt by association approach’ i.e. if the lncRNA share similar expression profile with known protein coding genes, the function of the lncRNA is probably similar to the function of the protein coding genes. In some cases, lncRNAs directly regulate the expression of neighbouring genes so the characterisation of the lncRNA genomic location and the expression of surrounding genes is essential.
To gain insight into the lncRNA role, we take advantage of publicly available data such as transcriptome data from the ENCODE Consortium to assess if our candidate lncRNAs are also expressed in other cell types or tissues or if their cellular localization (nucleus/cytoplasm) are known.
In addition, we are also interested in the upstream mechanism controlling the activation or suppression of the lncRNA expression. Again, we can take advantage of published data, such as ChIP-Seq if available, to assess the epigenetic regulation (chromatin marks) of the lncRNA gene or its transcription control by transcription factors.
Principal Investigator, Co-Investigators, Other researchers
Researchers from Andy’s Baker group
Andy Baker, Julie Rodor, Matt Bennett, Fatma Kok, Axelle Caudrillier, Joe Monteiro, Lin Deng, John Hung, Margaret Ballantyne
Collaboration within the University
David Newby, Jakub Kaczynsku, Alan Archibald, Lel Eory
External collaboration
Igor Ulitsky (Israel)

