SYCP1 head-to-head assembly is required for chromosome synapsis in mouse meiosis
Davies Lab – Science Advances
Authors
Billmyre, K.K., Kesler, E.A., Tsuchiya, D., Corbin, T.J., Weaver, K., Moran, A., Yu, Z., Adams, L., Delventhal, K., Durnin, M., Davies, O.R.* & Hawley, R.S.*
Summary of Paper by Jenna Hare
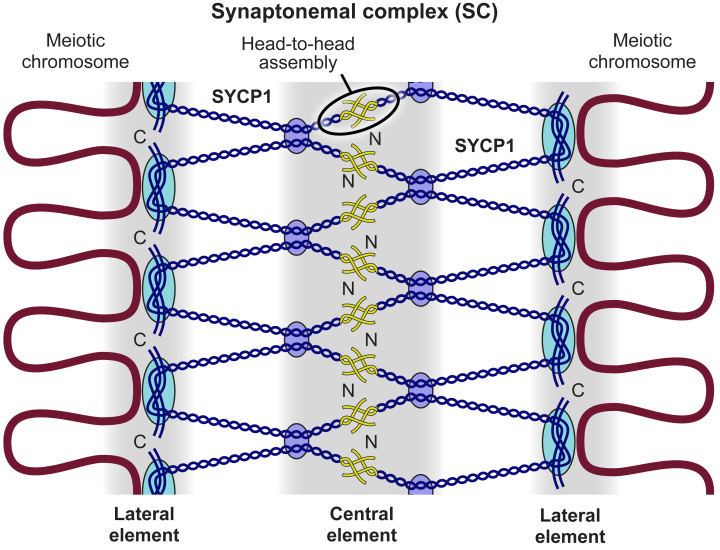
Meiosis is a biological process needed for successful sexual reproduction. During meiosis, homologous chromosome pairs line up together in a process called synapsis. After this, they segregate evenly to produce two haploid gamete cells ie. sperm and egg cells. Errors in meiosis can lead to aneuploidy, where there is an abnormal number of chromosomes in resulting sex cells. This can cause birth defects or infertility. A protein complex called the synaptonemal complex (SC) is vital for synapsis. Therefore, the scientists from The Stowers Institute and Wellcome Center for Cell Biology were interested in the components of the SC and how they interact together. They focused on a component called SYCP1. The two ends of a protein are named the N- and C-terminus, respectively. SYCP1 proteins arrange themselves head-to-head, with the N-termini facing each other. These scientists generated mice that have two mutations in the N-terminus of SYCP1 using Crispr-Cas9 technology. They could do this as SYCP1 is highly conserved between mice and humans. The mutations they created were called L102E and L106E. By combining their previous X-ray crystal structure with molecular dynamics simulations, they predicted that the mutation L106E would affect the head-to-head arrangement of SYCP1, whereas L102E would not. They used these models to test the importance of SYCP1 self-arrangement for successful synapsis. Using a range of techniques including histology, imaging of meiotic chromosomes, and fertility tests, they showed that the mutation L102E has little-to-no effect on synapsis, whereas the mutation L106E caused synapsis to be lost. A few key findings included how a modeled L102E protein could remain very stable, whereas a modeled L106E protein was unstable and disassociated. Furthermore, during chromosome alignment, L102E mice had similar distance between chromosomes to normal mice, whereas L106E mice had around triple the distance between them. These findings suggest that synapsis did not occur as the chromosomes are not properly aligned close to each other. Furthermore, fertility results showed that L106E mice had less mature sperm and therefore were infertile. Overall, they concluded that due to these findings, head-to-head self-arrangement of SYCP1 is vital for meiosis to occur. This research is useful to understand underlying reasons for infertility and birth defects in humans.

