Personal profile
- 2015 - present: Chancellor’s Fellow/Group Leader, University of Edinburgh
- 2011 - 2014: Senior Researcher, Norwegian Centre for movement disorders (NKB), Stavanger University Hospital, Norway
- 2007 - 2011: Postdoctoral researcher, Columbia University Medical Centre, New York, USA
- 2005 - 2007: EMBO Long-Term Fellow, Columbia University Medical Centre, New York, USA
- 2002 - 2005: Ph.D. in Developmental Biology, Max Planck Institute for Biophysical Chemistry, Göttingen, Germany
- 2000 - 2001: M.Sc. in Biotechnology, Wageningen University, The Netherlands
- 1999: Diploma in Biology and Biotechnology, Agricultural University of Athens, Greece
Research Theme
Research
1. The microbiota in Parkinson’s disease
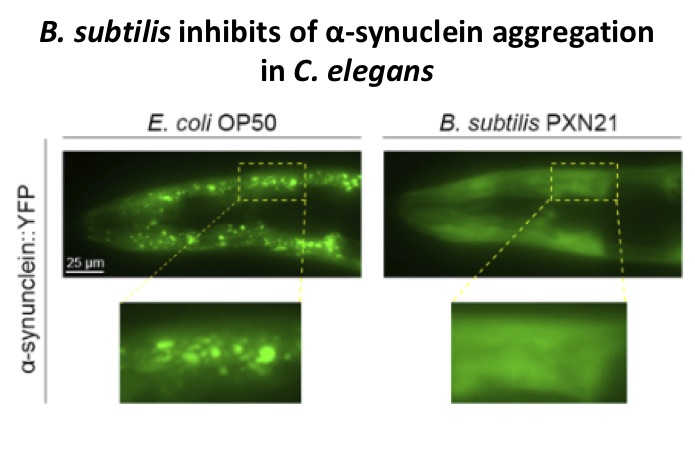
Parkinson’s disease is a progressive neurodegenerative disorder characterized by loss of dopaminergic neurons and alpha-synuclein aggregation. Recent studies have revealed a role for the gut microbiome in the development or progression of Parkinson’s. However, the complexity of the microbial ecosystem in the human gut makes it challenging to understand the contributions of individual species or the mechanisms by which they affect host physiology. The bacterivore C. elegans allows precise control of its microbiota and investigations at the single species-single gene level.

We aim to understand bacteria-host interactions and how they influence metabolic pathways, proteostasis, and neuronal integrity. We investigate bacteria present in the gut of Parkinson’s patients at different abundances that in healthy controls for their effects in C. elegans models of Parkinson’s disease.
2. Mechanisms of Bacillus subtilis protection against alpha-synuclein aggregation
Probiotic bacteria positively influence gastrointestinal function and also, through molecular communication networks, affect remote tissues and the brain. Our lab investigates the effects of probiotic bacteria present in over-the-counter products using C. elegans models of neurodegenerative disease.
We discovered that the probiotic bacterium B. subtilis inhibits human alpha-synuclein aggregation. We have linked this effect to a number of bacterial pathways and molecular changes in the host, including biofilm formation in the gut, the insulin signalling pathway and the sphingolipid metabolism pathway.
We are taking an inter-species multidisciplinary approach to understand the precise metabolic changes triggered in the host by the probiotic B. subtilis and identify the bacterial signals underlying the protection against alpha-synuclein aggregation. We are exploring the conservation of these findings in mice and in cellular models of Parkinson’s disease.
3. Mechanisms of dopaminergic neurodegeneration
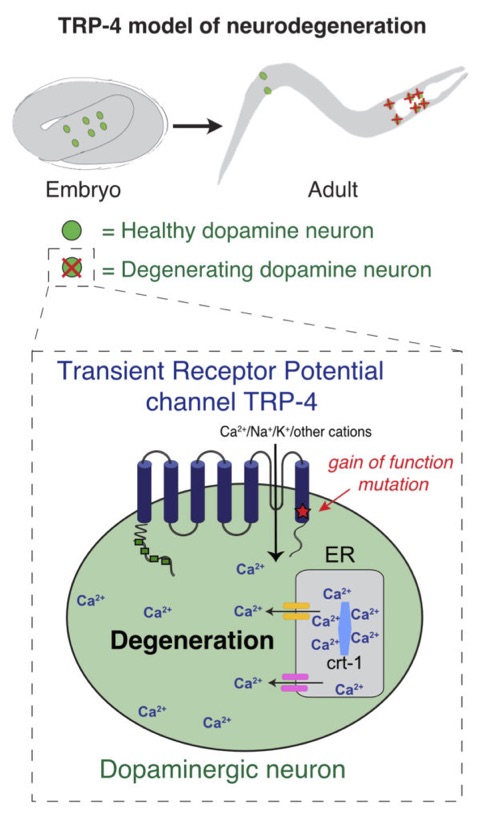
We are interested in the molecular mechanisms that safeguard the structural and functional integrity of dopaminergic neurons throughout life.
We have identified several mutations that impact the specification or survival of dopaminergic neurons. One of these mutations, a gain-of-function mutation in a Transient Receptor Potential channel, trp-4, causes dopamine neurons to progressively degenerate, through disrupting calcium homeostasis of the neuronal cell. Making use of the trp-4 model and high-throughput molecular genetics, we are investigating the cellular pathways that lead to neuronal cell death.
Funding
Team members
- Maria Doitsidou (PI)
- Feng Xue (Postdoc)
- Eugenia Goya (Postdoc)
- Lourdes Riquelme-Dominguez (PhD student)
- Janet Poplawski (PhD student, primary supervisor Prof. Kathryn Ball)
Previous lab members
- Viktoria Bajuszova (Research Assistant)
- Cristina Sampedro Torres-Quevedo (Masters student, Research assistant)
- Ailish Tynan (Masters student)
- Katy Dewison (Masters student)
Collaborations
- Dr. Emanuel Busch, CDBS, University of Edinburgh
- Dr. Sebastian Greiss, CDBS, University of Edinburgh
- Dr. David Breen, CCBS, University of Edinburgh
- Dr. Jodi Grødem, Stavanger University Hospital, Norway
- Dr. Louise Horsfall, University of Edinburgh
- Dr. Tilo Kunath, University of Edinburgh
- Dr. Claire Benard, UQam
- Prof. Oliver Hobert, Columbia University Medical Centre, New York
Selected publications
- Romanelli-Credrez L, Doitsidou M, Alkema MJ, Salinas G. HIF-1 Has a Central Role in Caenorhabditis elegans Organismal Response to Selenium. Front Genet. 2020 Feb 25;11:63
- Goya, ME, Xue, F*, Sampedro-Torres-Quevedo*, C, Arnaouteli, S, Riquelme-Dominguez, L, Romanowski, A, Brydon, J, Ball, K, Stanley-Wall, NR, and Doitsidou, M§. Bacillus subtilis prevents and reverses α-synuclein aggregation in C. elegans. Cell Reports. 2020 Jan 14;30(2):367-380
- Doitsidou M§, Minevich G, Kroll JR, Soete G, Gowtham S, Korswagen HC, Sebastiaan van Zon J, Hobert O. A Caenorhabditis elegans Zinc Finger Transcription Factor, ztf-6, Required for the Specification of a Dopamine Neuron-Producing Lineage. G3 (Bethesda). 2018 Jan 4;8(1):17-26
- Katsanos D, Koneru SL, Mestek Boukhibar L, Gritti N, Ghose R, Appleford PJ, Doitsidou M, Woollard A, van Zon JS, Poole RJ, Barkoulas M. Stochastic loss and gain of symmetric divisions in the C. elegans epidermis perturbs robustness of stem cell number. PLoS Biol. 2017 Nov 6;15(11):e2002429
- Doitsidou, M§, Jarriault, S§. & Poole, RJ§. Next-generation sequencing-based approaches for mutation mapping and identification in C. elegans. 1 Oct 2016 (Wormbook methods) Genetics. 204, 2, p. 451-474
- Bénard, C. & Doitsidou, M§. Nervous System Ageing. 7 Dec 2016. Springer, p. 163-189 10.1007/978-3-319-44703-2_82016
- Nagarajan A, Ning Y, Reisner K, Larsen JP, Hobert O*, Doitsidou M*§.Progressive degeneration of dopaminergic neurons through TRP channel-induced cell death. J Neuroscience. 2014 Apr 23;34(17): 5738-46
- Doitsidou M*§, Flames N*§, Topalidou N, Abe N, Felton T, Remesal L, Popovitchenko T, Mann R, Chalfie M and Hobert O§. A combinatorial regulatory signature controls terminal differentiation of the dopaminergic nervous system in C. elegans. Genes and Development. 2013 Jun 15;27(12):1391-405
- Doitsidou M§, Poole JR, Sarin S, Bigelow H and Hobert O§. C. elegans mutant identification with a one-step Whole-Genome-Sequencing and SNP mapping strategy. PLoS ONE. 2010 Nov 8;5(11):e15435.
- Bigelow H, Doitsidou M, Sarin S, Hobert O. MAQGene: software to facilitate C. elegans mutant genome sequence analysis. Nature Methods. 2009 Aug;6(8):549.
- Doitsidou M§, Flames N, Lee AC, Boyanov A, Hobert O. Automated screening for mutants affecting dopaminergic neuron specification in C. elegans. Nature Methods. 2008 Oct;5(10):869-72.
- Thorpe JL*, Doitsidou M*, Ho SY, Raz E and Farber SA. Germ Cell Migration in Zebrafish Is Dependent on HMGCoA Reductase Activity and Prenylation. Developmental Cell. 2004 Feb;6(2):295-302.
- Doitsidou M, Reichman-Fried M, Stebler J, Köprunner M, Dörries J, Meyer D, Esguerra VC, Leung T and Raz E. Guidance of primordial germ cell migration by the chemokine SDF-1. Cell 2002 Nov 27;111(5):647-59.
§: corresponding author, *: equally contributing
Information for students:
Willingness to discuss research projects with undergraduate and postgraduate students: YES - please click here



