Study sheds light on bacterial sharing between humans and animals
A recent paper demonstrates how infectious diseases and antimicrobial resistance can be shared between humans and animals in an urban environment.
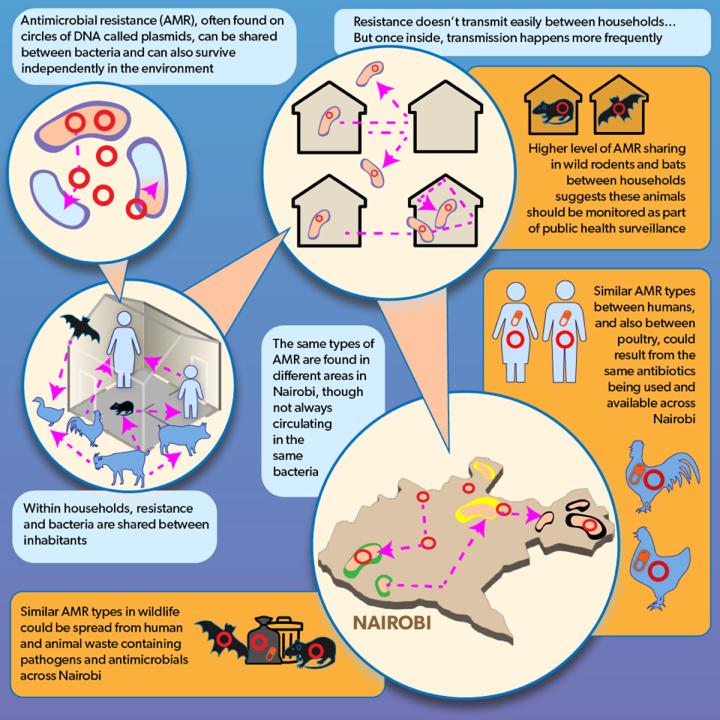
The study was conducted by researchers at the University of Edinburgh, the International Livestock Research Institute (ILRI), Nairobi, Kenya, and the University of Liverpool, as part of a project investigating the impact of urbanisation on pathogen emergence and transmission.
The authors sampled a range of humans, animals (both livestock and wildlife) in different parts of Nairobi, Kenya, to help understand how bacteria are shared between hosts sharing the same household environment.
Bacterial transmission within and between species
They examined the distribution of a common and widespread species of bacteria, Escherichia coli, that is found in the guts of mammals and birds, and which can also cause serious illness in humans.
Using high-resolution genome sequencing technologies to examine the bacteria carried by household residents, they found that while most bacterial sharing occurs within the same host species living together in the same household – for instance human-to-human transmission – bacteria are also shared between humans, livestock and wildlife. The same strain of bacteria present in a human could also be found in a chicken.
Implications for antimicrobial resistance
This inter-species transmission has important implications for the spread of resistance to antibiotics.
Increases in antimicrobial resistance have been seen in many developing countries, fuelled in part by inappropriate use of antibiotics in both human and animals. The sharing of bacteria between hosts within a domestic setting demonstrates the potential for antimicrobial-resistant bacteria that emerge in one organism to move into others.
Understanding global spread
This study showcases the power of sequencing technologies combined with an epidemiologically structured sampling framework, to help the understanding of the spread of infectious diseases between humans and the animals that live around them.
The work also highlights the need for a global approach to address these issues.
Ultimately, the lessons learned in the project have implications not just for the developing nations: ‘The E. coli in Nairobi represents the E. coli across the entire world. What is being seen in Nairobi today could easily be in New York or Paris by tomorrow morning.’
Understanding more about transmission of microbes between livestock and humans is crucial to protecting health, globally. This paper is an excellent example of work co-led by The Universities of Edinburgh and Liverpool as part of a wide collaboration, working with the team in Nairobi to offer insights of global relevance. The rapid development of the city along with practice of keeping livestock within urban households has offered a powerful setting to develop our understanding of how antimicrobial resistance arises.
Read the paper in Nature Microbiology


