Identifying host genes important for human cytomegalovirus replication using siRNA screens
The Grey lab uses a number of screens which utilise of small interfering RNA (siRNA) to silence cellular genes in order to identify those which play a role in human cytomegalovirus (HCMV) replication.
Disease outcome from virus infection depends on complex host-pathogen interactions. Genome wide siRNA screens have proven a useful tool for deciphering which cellular genes are required for virus replication and which genes have antiviral properties. As well as contributing to our understanding of virus-host interactions, siRNA screens are a powerful approach for the identification of antiviral drug targets. The majority of screens performed measure virus replication through the expression of a reporter construct. Although this identifies entry and replication phenotypes, the crucial parameter of virus production is not considered.
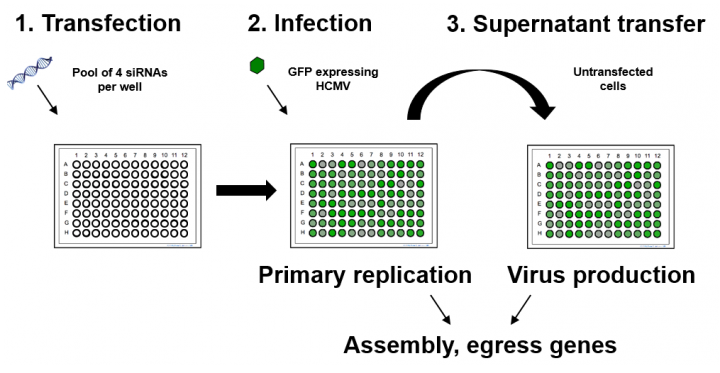
We have devised a novel siRNA screen that measures both primary replication but crucially, also measures infectious progeny. We have shown that the correlation between primary replication and infectious progeny is far lower than would be expected, indicating that measurement of primary replication alone may be misleading and the two-step approach is therefore superior to conventional one step approaches. Using this approach, we have identified multiple host factors critical to HCMV replication and virion production. Characterisation of these interactions has contributed to our knowledge of how the virus replicates, how cellular pathways contribute to HCMV replication and have demonstrated the effectiveness of this approach for the identification of novel small molecule inhibitors with potential as HCMV antivirals.
Role of Ubiquitin signalling in HCMV replication
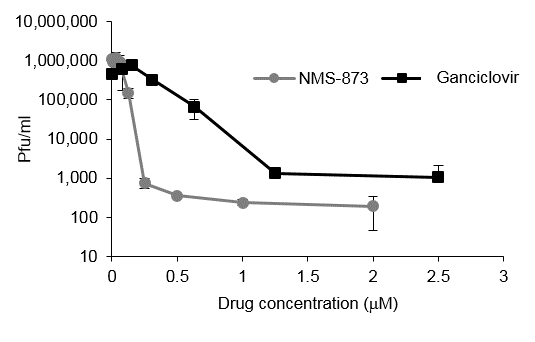
Previously, we identified the host gene ATP6V0C as a critical factor in HCMV virus production. ATP6V0C is a component of the vacuolar ATPase, and among other functions, is involved in membrane organization. To identify additional host membrane organization factors involved in HCMV replication we performed a focused siRNA screen against 160 host genes involved in membrane organization using pools of four siRNAs against each target. One of the hits that caused the greatest attenuation of HCMV replication was Valosin containing protein (VCP) a member of the hexameric AAA ATPase family that plays a pivotal role in ubiquitin mediated signaling through remodelling and degradation of target proteins. We showed that knockdown of the host ubiquitin-dependent segregase VCP/p97, results in loss of IE2 expression, subsequent suppression of early and late gene expression and, ultimately, failure in virus replication. The human cytomegalovirus major immediate early proteins IE1 and IE2 are critical drivers of virus replication and are considered pivotal in determining the balance between productive and latent infection. VCP has been identified as a potential anti-cancer target with highly specific and potent small molecule inhibits such as NMS-873 available. We show that NMS-873, a small molecule inhibitor of VCP, is a potent HCMV antiviral with potential as a novel host targeting therapeutic for HCMV infection. This study demonstrated the effectiveness of the approach for understanding critical cellular pathways for HCMV replication and the identification of potential novel antiviral therapies. We are currently investigating the targets of VCP during HCMV replication and investigating ubiquitin signalling in HCMV replication on a more global scale.
Identification of host factors important for viral assembly and egress
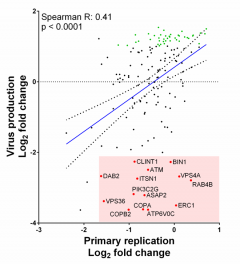
As obligate intracellular parasites, viruses are completely dependent on host factors for replication. Assembly and egress of complex virus particles, such as human cytomegalovirus, is likely to require many host factors. Despite this, relatively few have been identified and characterised. Previously, we identified the host gene ATP6V0C as a critical factor in HCMV replication. Knockdown of ATP6V0C resulted in modest inhibition of virus replication (as defined by virus gene expression and DNA amplification), but a pronounced reduction in virus progeny, suggesting a defect in assembly or egress. ATP6V0C is a component of the vacuolar ATPase, and among other functions, is involved in membrane organisation. Using our two-step screening approach and a siRNA library against 160 host genes involved in membrane organisation we identified multiple host factors, including ERC1, RAB4B, COPA, and COPB2, which caused profound loss of virus production despite only minor effects on virus replication. We are currently characterising the functional role of these proteins in the assembly and egress of HCMV.
Current and future studies
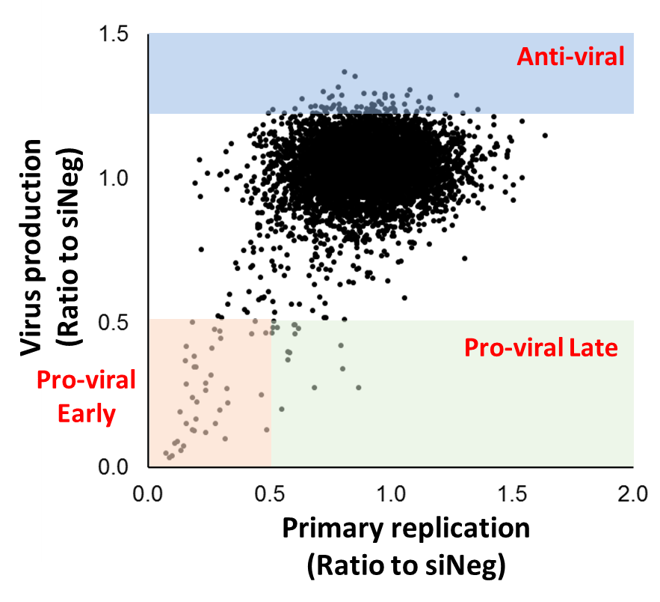
We have now expanded our two-step approach to include our largest siRNA screen to date, using a library of nearly 8000 siRNAs against druggable host genes. To our knowledge this is the largest siRNA screen against HCMV. This screen has resulted in identification of multiple host genes important for HCMV replication and virus production, including related genes within multiprotein complexes and cellular pathways. Studies are ongoing to characterise these hits and corresponding small molecule inhibitors.

