Diagnosis of active trypanosomiasis through a parasite specific small RNA
The Grey lab and our collaborators in the Morrison lab have identified a parasite specific 7SL-derived small RNA for the diagnosis of active trypanosomiasis infection
Human and animal African trypanosomiasis (HAT & AAT, respectively) remain a significant health and economic issue across much of sub-Saharan Africa. Effective control of AAT and potential eradication of HAT requires affordable, sensitive and specific diagnostic tests that can be used in the field. Our findings identify a species-specific trypanosome small RNA that can be detected at high levels in the serum of cattle with active parasite infections. This provides the basis for the development of a cheap, non-invasive and highly effective diagnostic test for trypanosomiasis.
African trypanosomes, vector borne protozoa transmitted by tsetse flies (Glossina species), cause HAT and AAT across sub-Saharan Africa. AAT, caused by Trypanosoma congolense, Trypanosoma vivax and Trypanosoma brucei affects approximately 70 million cattle and kills 3 million cattle per year, and is one of the most significant infectious disease constraints upon agriculture in the region. HAT is caused by two variants of T. brucei, T. b. gambiense and T. b. rhodesiense, and in recent years the impact of this disease has been significantly reduced through active case detection, with <3,000 cases reported in 2015, down from ~50,000 in 2000. However, new and improved tools are required for both diseases; for AAT as a tool to begin to reduce the current significant burden, and for HAT to facilitate the delivery of the World Health Organisation (WHO) aim of HAT elimination by 2030. The ability to diagnose active infections is currently still a significant challenge for both AAT and HAT.
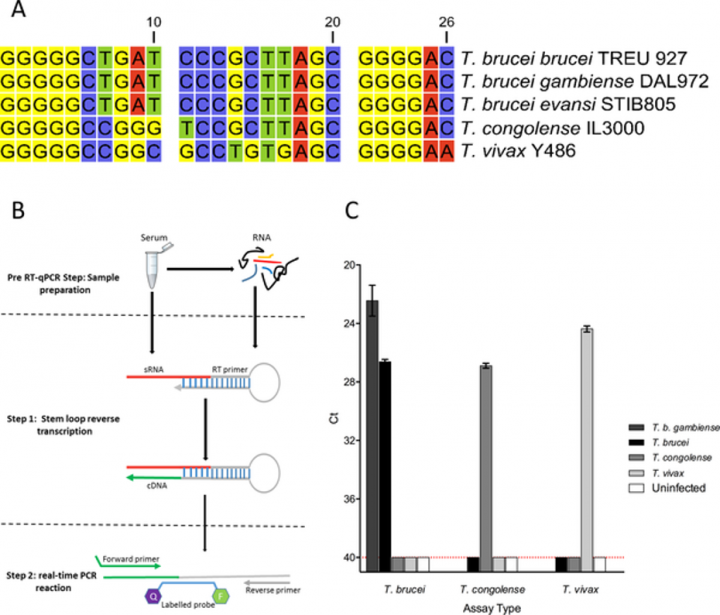
There is therefore a significant unmet need for a pathogen marker of active infection that accurately indicates whether an animal or human is currently infected. Through analysing the blood of cattle infected with trypanosomes, we identified a short sequence of RNA that was present at very high levels. This small RNA derives from the trypanosome genome, and we could identify its presence in the genome of all three species that are responsible for human and animal disease. We were able to design species-specific tests, and showed that in samples from infected animals the assays were more sensitive than the traditional microscope-based detection, importantly the signal disappeared quickly after successful treatment, and when treatment failed, the assay was able to accurately identify when infection persisted. We also demonstrated that the causative agent of human trypanoaomiasis secretes the marker at similar levels to that seen in the animal-infective trypanosomes. Therefore, we have discovered a marker of trypanosome infection that is present at high levels in the blood of infected animals, disappears quickly upon successful treatment, but is effective at detecting instances of unsuccessful treatment and persistent infection. This represents a potentially powerful diagnostic tool for human and animal trypanosomiasis.

