Michlewski Lab publish a collaborative paper in Molecular Cell
An international group of researchers led by Dr Alfredo Castello (University of Oxford) developed ‘comparative RNA-interactome capture’ to analyse the RNA-bound proteome during virus infection. >200 cellular RNA-binding proteins change their binding activity in response to this challenge, mainly driven by transcript availability. Many of these RNA-binding proteins regulate viral replication and can be targeted to influence infection outcome.
Publication summary
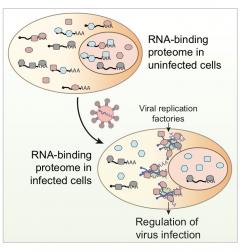
The compendium of RNA-binding proteins (RBPs) has been greatly expanded by the development of RNA-interactome capture (RIC). However, it remained unknown if the complement of RBPs changes in response to environmental perturbations and whether these rearrangements are important. To answer these questions, we developed “comparative RIC” and applied it to cells challenged with an RNA virus called sindbis (SINV). Over 200 RBPs display differential interaction with RNA upon SINV infection. These alterations are mainly driven by the loss of cellular mRNAs and the emergence of viral RNA. RBPs stimulated by the infection redistribute to viral replication factories and regulate the capacity of the virus to infect. For example, ablation of XRN1 causes cells to be refractory to SINV, while GEMIN5 moonlights as a regulator of SINV gene expression. In summary, RNA availability controls RBP localization and function in SINV-infected cells.
Link to publication

