Using single molecule sequencing to understand the regulation of DNA methylation
Dr Duncan Sproul & Professor Ramon Grima
About the Project
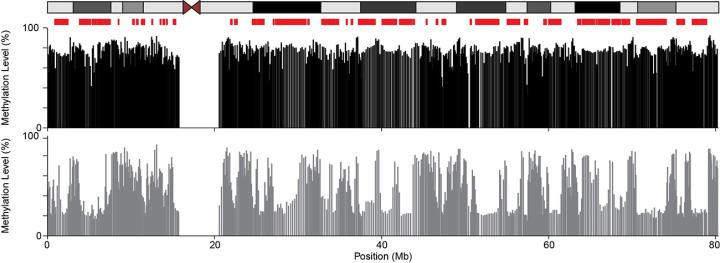
DNA methylation is a repressive epigenetic mark whose genomic pattern is established early in mammalian development. This process must be completed correctly for normal development to proceed. In addition, disruption of developmentally established methylation patterns is associated with aging and disease. By defining genome-wide DNA methylation patterns, high throughput sequencing has provided invaluable insight into the mechanisms that underpin the establishment and maintenance of DNA methylation in the genome (1). However, this has primarily focused on the analysis of bulk population sequencing which provides limited information about the underlying dynamics generating steady-state methylation patterns. We have recently demonstrated that Nanopore sequencing can be used to probe DNA methylation at the single molecule level genome-wide (2).
This PhD project focuses on using this approach to dissect the contribution that different DNA methyltransferases make to DNA methylation patterns in different parts of human genome. During the PhD, the student will analyse knockout cells lacking different DNA methyltransferase enzymes by Nanopore sequencing to understand their contribution to DNA methylation dynamics at the single molecule level. They will then go further dissect this effect using mutations of key domains and mathematical modelling of the data.
The project is an interdisciplinary collaboration between the Sproul and Grima groups at the University of Edinburgh’s MRC Human Genetics Unit and School of Biological Sciences. The student will be trained in the generation and analysis of cutting-edge high-throughput sequencing data drawing on expertise of both groups through laboratory experiments, bioinformatic analyses and computational modelling to understand how DNA methyltransferases regulate DNA methylation patterns in vivo.
The project would ideally suit those with a quantitative background in biology or another discipline who have an interest applying this training to understand epigenetic regulation. Applicants with other training backgrounds who are motivated to learn the necessary computational skills will also be considered.
Funding information and application procedures:
This 4 year PhD project is part of a competition funded by EASTBIO BBSRC Doctoral Training Partnership (DTP).
EASTBIO Application and Reference Forms can be downloaded via http://www.eastscotbiodtp.ac.uk/how-apply-0
Please send your completed EASTBIO Application Form along with a copy of your academic transcripts to student-admin@igc.ed.ac.uk
You should also ensure that two references have been sent to student-admin@igc.ed.ac.uk by the deadline using the EASTBIO Reference Form.
This opportunity is open to UK and international students and provides funding covering stipend and UK level tuition fees. The University of Edinburgh covers the difference between home and international fees meaning that the EASTBIO DTP offers fully-funded studentships to all appointees. There is a cap on the number of international students the DTP recruits. It is therefore important for us to know from the outset which fees status category applicants will fall under when applying to our university.
Please refer to UKRI website and Annex B of the UKRI Training Grant Terms and Conditions for full eligibility criteria.


