Dr Chih-Jen (Lance) Lin
Research interests, biography and patents.
Dr Chih-Jen (Lance) Lin
Lecturer

- Principal Investigator
- Royal Society of Edinburgh Personal Research Fellow funded by the Scottish Government
Contact details
- Email: Chih-Jen.Lin@ed.ac.uk
Address
- Street
The Centre for Reproductive Health
Institute for Regeneration and Repair (IRR)
4-5 Little France Drive
Edinburgh BioQuarter- City
- Edinburgh
- Post Code
- EH16 4UU
Research Interests
The oocyte, the female gamete, holds the remarkable capacity to reprogramme cells into a totipotent embryo which can generate all types of cells in an organism. Fertilisation is the process of combining male and female gametes, which triggers extensive reprogramming events such as protamine-to-histone exchange, remodelling of histones, DNA demethylation and initiation of transcription in embryos. Genetic and epigenetic abnormalities arising during developmental processes lead to failure or birth defects. Approximately one in seven couples, ~3.5 million people in the UK, have difficulty conceiving (defined as infertility). Dr. Chih-Jen Lin’s research focuses on female infertility specifically in oocyte health, fertilisation and early embryogenesis.
Dr. Lin’s previous studies have demonstrated that epigenetic regulators such as histone variant H3.3 and transcription of ribosomal RNAs have pivotal roles in fertilisation (Figure A) and pre-implantation embryonic development (Figure B) in mouse models.
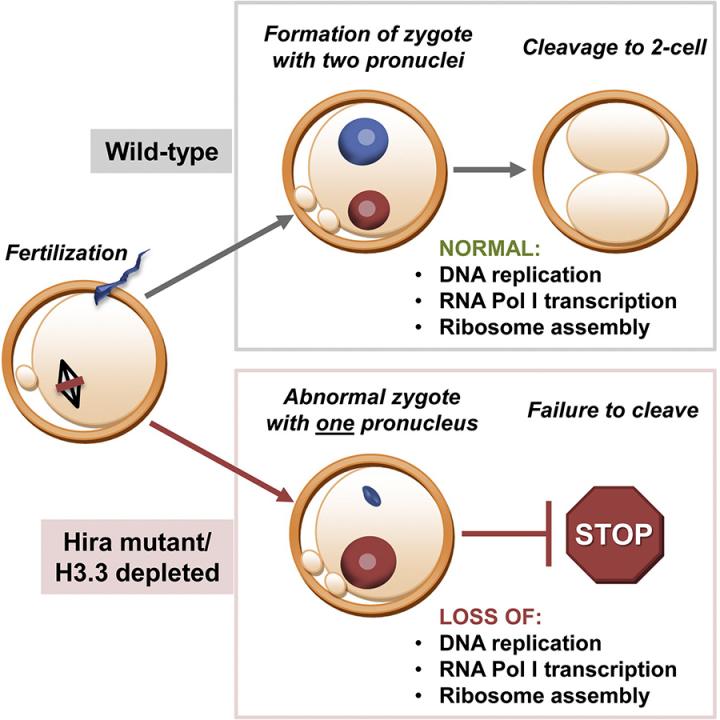
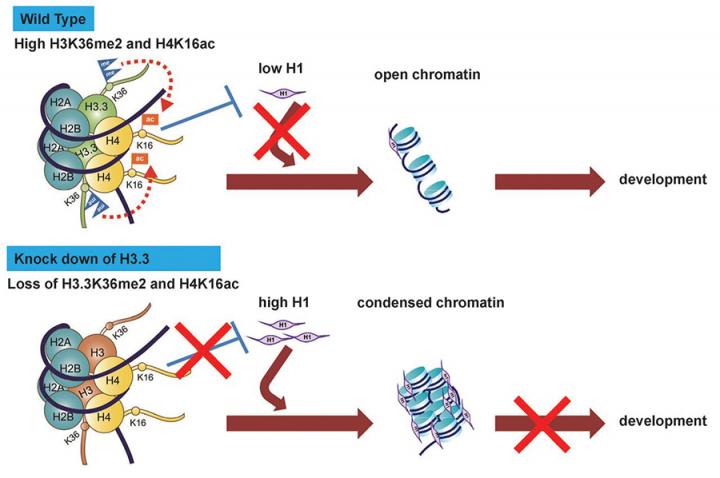
Dr. Lin’s long term research goals are to understand epigenetic regulation during oocyte-to-embryo transition and how it impacts on female infertility.
Recent research outcomes:
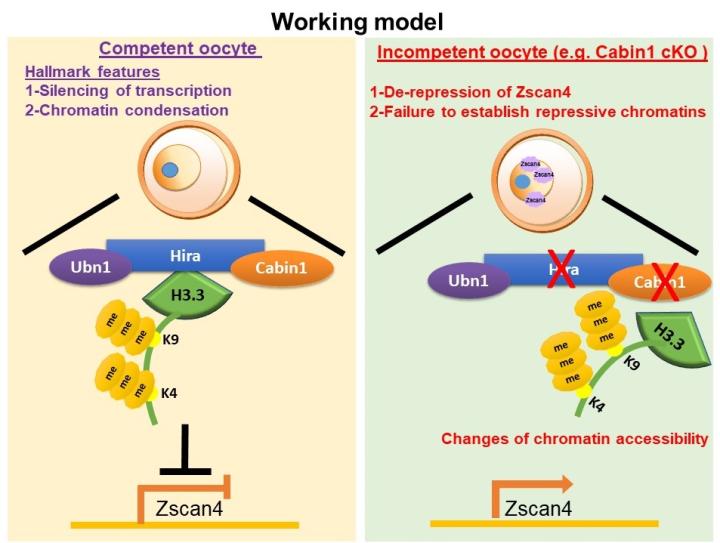
- The developmental and molecular roles of histone variant, H3.3, and its chaperones during oocyte-to-embryo transition.
We have generated 3 new mouse models of female infertility (Hira and Cabin1 conditional mutants and Ubn1 knockdown oocytes). We showed that H3.3 and its chaperones are involved as ‘key repressors’ in establishing condensed chromatin status and so regulating quiescent transcription status during the final stage of oocyte growth (Figure C; Smith et al., 2022 Development).
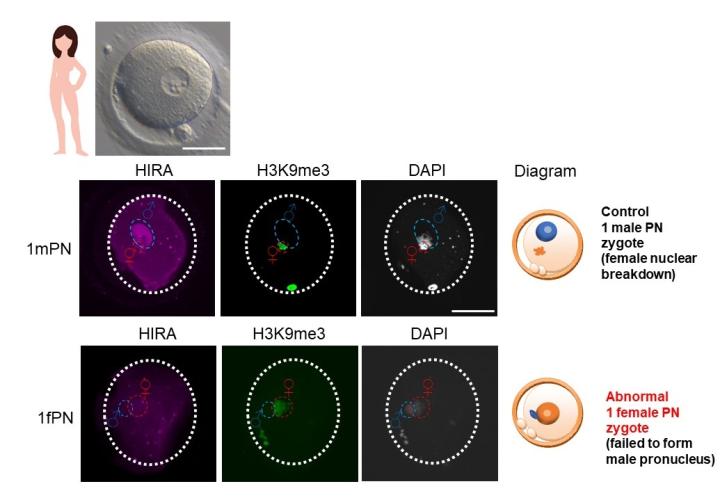
- Investigation of the molecular mechanism of human abnormal one-pronucleus zygotes after IVF treatment.
We have been awarded an HFEA research licence to use human embryos for investigating the underlying mechanism of abnormal fertilisation. We provided evidence that the abnormal one-pronucleus phenotype which we observed in mice (loss-of-function in Hira, Cabin1, and Ubn1) is likely conserved in humans (Figure D; Smith et al., 2021 Reproduction).
- Genome-wide translatomic and functional analyses of totipotent embryos.
We have recently demonstrated that ribosomal RNAs (Lin et al., 2014 Dev Cell) and repetitive long non-coding RNAs, Line1 (Percharde, Lin et al., 2018 Cell; Percharde, Lin et al., 2021 STAR Protocols), are essential for the initiation of ZGA and the establishment of totipotent embryos in mice. To further investigate this process, we have developed a new ultra-low input Polysomal RNA isolation protocol for RNA-sequencing (Masek et al., 2020 Int J Mol Sci) and generated novel datasets of the embryos by isolation of the actively-translating molecules.
Biography
Chih-Jen (Lance) Lin received his BS and MS degree from the National Taiwan University in Taiwan working on animal transgenesis in the lab of the prestigious embryologist Prof. Winston Teng-Kuei Cheng (renowned as the world’s first scientist to produce test-tube pigs and sheep). He obtained his PhD degree from University of Connecticut, USA under the supervision of Animal Cloning experts: Prof. Jerry (Xiangzhong) Yang and Cindy (Xiuchun) Tian. He received his solid postdoctoral training of stem cell biology/epigenetics (Dr. Miguel Ramalho-Santos Lab) and oocyte biology (Prof. Marco Conti Lab) in University of California San Francisco, USA. He then conducted a short second postdoctoral training in Prof. Philippe Soriano’s Lab in Icahn School of Medicine at Mount Sinai, New York, USA.
In 2015, He was recruited to the MRC Centre for Reproductive Health at the University of Edinburgh as a Senior Research Fellow to establish his own research group and awarded a Royal Society of Edinburgh Personal Research Fellowship in 2016.
Current research projects
The developmental and molecular roles of histone variant, H3.3, and its chaperones during oocyte-to-embryo transition.
We have generated 3 new mouse models of female infertility (Hira and Cabin1 conditional mutants and Ubn1 knockdown oocytes). We showed that H3.3 and its chaperones are involved as ‘key repressors’ in establishing condensed chromatin status and so regulating quiescent transcription status during the final stage of oocyte growth (Figure C; Smith et al., 2022 Development).
Investigation of the molecular mechanism of human abnormal one-pronucleus zygotes after IVF treatment.
We have been awarded an HFEA research licence to use human embryos for investigating the underlying mechanism of abnormal fertilisation. We provided evidence that the abnormal one-pronucleus phenotype which we observed in mice (loss-of-function in Hira, Cabin1, and Ubn1) is likely conserved in humans (Figure D; Smith et al., 2021 Reproduction).
Genome-wide translatomic and functional analyses of totipotent embryos.
We have recently demonstrated that ribosomal RNAs (Lin et al., 2014 Dev Cell) and repetitive long non-coding RNAs, Line1 (Percharde, Lin et al., 2018 Cell; Percharde, Lin et al., 2021 STAR Protocols), are essential for the initiation of ZGA and the establishment of totipotent embryos in mice. To further investigate this process, we have developed a new ultra-low input Polysomal RNA isolation protocol for RNA-sequencing (Masek et al., 2020 Int J Mol Sci) and generated novel datasets of the embryos by isolation of the actively-translating molecules.
Staff / group members
Ms Rowena Smith
Current PhD/MD students
Heather Flanagan (co-supervised)
Current and recent grants
2018:
- MRC Flexible Supplement Fund for the PhD student.
- RSE International Exchange Programme (Partner: Ministry of Science and Technology, Taiwan; National Taiwan University, Prof Li-Ying Sung).
- The Barbour Watson Trust
- University of Edinburgh CMVM College Fund
2017:
- RSE International Exchange Programme (Partner: Academy of Sciences of the Czech Republic; Dr Andrej Susor).
- MRC Flexible Supplement Fund for the PhD student.
- University of Edinburgh Moray Endowment Fund.
- The Barbour Watson Trust for the lab member.
- SRF Vacation Scholarship for the lab member.
2016-2021:
Royal Society of Edinburgh Personal Research Fellowship;” Investigating the underlying mechanism and the development of a novel rescue strategy for abnormally developing zygotes.”.
2015:
Wellcome Trust-University of Edinburgh Institutional Strategic Support Fund; “Investigation the roles of histone variant H3.3 and incorporation mechanisms during oocyte-to-embryo transition.”.
Selected recent publications
Smith R, Z Jiang, A Susor, H Ming, J Tait, M Conti, and CJ Lin. The H3.3 chaperone Hira complex orchestrates oocyte developmental competence. 2022 Development 149 (5): dev200044.
Percharde, M, Lin CJ, and Ramalho-Santos M. Depletion of Nuclear LINE1 RNA in Mouse ESCs and Embryos. STAR Protocols 2021; 2(3) 100726.
Smith R, Pickering S, Kopakaki, A, Thong, KJ, Anderson, RA and Lin CJ. Histone H3.3 Hira chaperone complex contributes to zygote formation in mice and is implicated in human 1PN zygote phenotype. Reproduction 2021; 161(6): 697-707.
Aleshkina D, Lyyappan R., Lin CJ, Masek T, Pospisek M, and Susor A. ncRNA BC1 influences translation in the oocyte. RNA Biol. 2021 ; 18(11) :1893-1904.
Masek T, E del Llano, L Gahurova, M Kubelka, A Susor, K Roucova, CJ Lin, AW Bruce, and M Pospisek. Identifying the translatome of mouse NEBD-stage oocytes via SSP-profiling- A novel polysomme fractionation method. Int J Mol Sci. 2020; 21(4):1254.
Percharde, M, Lin, CJ, Yin, Y, Guan, J, Peixoto, GA, Bulut-Karslioglu, A, Biechele, S, Huang, B, Shen, X, and Ramalho-Santos, M. A LINE1-Nucleolin Partnership Regulates Early Development and ESC Identity. Cell. 2018;174:391-405.
Fraser R, Lin CJ. Epigenetic reprogramming of the zygote in mice and men: on your marks, get set, go! Reproduction. 2016;152(6):R211-R22.
Guzman-Ayala M, Sachs M, Koh FM, Onodera C, Bulut-Karslioglu A, Lin CJ, et al. Chd1 is essential for the high transcriptional output and rapid growth of the mouse epiblast. Development. 2015;142(1):118-27.
Biechele S, Lin CJ, Rinaudo PF, Ramalho-Santos M. Unwind and transcribe: Chromatin reprogramming in the early mammalian embryo. Current opinion in genetics & development. 2015;34:17-23.
Lin CJ, Koh FM, Wong P, Conti M, Ramalho-Santos M. Hira-Mediated H3.3 Incorporation Is Required for DNA Replication and Ribosomal RNA Transcription in the Mouse Zygote. Dev Cell. 2014;30(3):268-79.
Lin CJ, Conti M, Ramalho-Santos M. Histone variant H3.3 maintains a decondensed chromatin state essential for mouse preimplantation development. Development. 2013;140(17):3624-34.
Chen J, Torcia S, Xie F, Lin CJ, Cakmak H, Franciosi F, et al. Somatic cells regulate maternal mRNA translation and developmental competence of mouse oocytes. Nat Cell Biol. 2013;15(12):1415-23.
Tang Y, Luo Y, Jiang ZL, Ma YH, Lin CJ, Kim C, et al. Jak/Stat3 Signaling Promotes Somatic Cell Reprogramming by Epigenetic Regulation. Stem Cells. 2012;30(12):2645-56.
Tang Y, Lin CJ, Tian XC. Functionality and Transduction Condition Evaluation of Recombinant Klf4 for Improved Reprogramming of iPS Cells. Cell Reprogram. 2011;13(2):99-112.
Lin CJ, Amano T, Zhang J, Chen YE, Tian XC. Acceptance of Embryonic Stem Cells by a Wide Developmental Range of Mouse Tetraploid Embryos. Biol Reprod. 2010;83(2):177-84 (Cover Story).
Patents
Cheng, WTK, CM Chen, SW Lin, CH Wang, CJ Lin, and SC Wu. 2004. Transgenic mammal secreting B-domain deleted human FVIII in its milk (USA Patent No. US 7,667,089 B2)
Cheng, WTK, CM Chen, SH Lin, CH Wang, CJ Lin, and SC Wu. 2007. Transgenic animals producing biologically active human factor VIII in their milk driven by mammary-specific expression cassette with intact or B domain-deleted recombinant hFVIII gene constructs. (ROC Patent, Oct 01, 2007~Feb 05, 2024; Patent number: ROC I-287,578.)
Lee, KH, HW Hang, HR Chang, CF Tu, and CJ Lin. 2004. A Method for generating non-human mammalian chimeric embryo. (ROC accepted on Mar 21th, 2010 (092136457) and USA Patents Pended, Pending No.10/875,225.)
Lee, KH, HW Hang, HR Chang, CF Tu, and CJ Lin. 2005. A Method for generating non-human mammalian chimeric embryo. (China Patent CN200410000197.5.)
Cheng WTK, SC Wu, CC Hsu, YS Lin, and CJ Lin. 2011. Novel porcine pancreatic amylase gene promoter and transgenic pigs expressing heterologous digestive enzymes. (USA Patent No. 7,956,238 B2)
Cheng WTK, SC Wu, CC Hsu, YS Lin, and CJ Lin. 2011. Novel porcine pancreatic amylase gene promoter and transgenic pigs expressing heterologous digestive enzymes. (ROC Patent, Oct 21, 2011~May 16, 2026; Patent number: TW095117405)
Technology transfer
Cheng, WTK, SC Wu, and CJ Lin. 2005. Transgenic animal system for mammary-gland-specific expression platform (To the TaiMont Biotechnology Ltd., Jan. 2005 ~ Jan. 2010).
Recent awards
2020: Basic Science Award for oral presentation in ESHRE annual meeting.
2014: Joy Cappel (Rockland) 2014 Young Investigator Award.
2014: Betty Jean Ogawa Memorial Awards for the top-scoring poster presentations in ISSCR Annual Meeting.
2014: Travel award to the ISSCR Annual Meeting Jun 2014.
Current external activities
Visiting Professor of University of Pavia, Italy

