Adhesion protein networks reviewed by CRUK EC scientists
Adhesion protein networks reviewed by CRUK EC scientists: 1 March 2016
For cells to coordinate complicated processes, like their migration through tissues, networks of proteins must work together. It is now clear that the protein machines that control such processes in cells are extremely complex.
In fact, research into the complex protein machinery that controls cell adhesion is revealing much more about the biology of cells – and diseases like cancer – than we had anticipated.
Networks of adhesion proteins and their interactions in cells
Cell adhesion is essential for multicellular life to exist.
Cell adhesion is mediated by adhesion receptors, which are proteins on the surface of cells. Adhesion receptors bind to the extracellular matrix – a complex mixture of proteins and sugars that surrounds cells, providing mechanical support and initiating biochemical signals in cells. Cells sense and integrate this extracellular information at cell-matrix contacts using adhesion receptors, such as integrins.
During cell adhesion, adhesion proteins inside cells are recruited to adhesion receptors to form multi-protein assemblies called adhesion complexes. The networks of adhesion proteins and their interactions work to control the migration, proliferation and survival of cells. When cell adhesion goes wrong, these processes malfunction, and disease often results. So, working out how cell adhesion systems function as interacting networks is currently under intense scrutiny.
This is the focus of a review article, published recently as an open-access article in Current Opinion in Cell Biology by Dr. Adam Byron and Prof. Margaret Frame from the Cancer Research UK Edinburgh Centre.
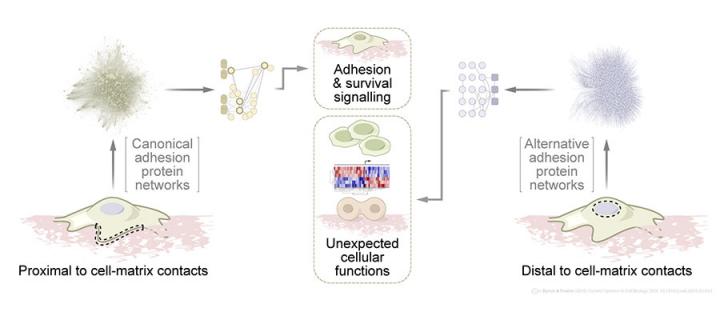
But adhesion proteins do not just spend their time near the plasma membrane in adhesion complexes. Recent research has uncovered wider roles for adhesion proteins away from adhesion receptors, and the article also highlights the important functions of adhesion proteins in diverse subcellular locations.
The multi-protein adhesion complexes that form when integrin adhesion receptors bind to extracellular proteins consist of scaffolding, adaptor and signalling proteins. These proteins connect integrins to the cell’s cytoskeleton, and they have been collectively termed the ‘adhesome’.
Until recently, the comprehensive analysis of adhesomes was restricted by the difficulties of isolating adhesion complexes from cells, as they are not very stable and they are connected to various parts of the cell, like the plasma membrane and the cytoskeleton. These difficulties have largely been overcome with the development of biochemical methods to purify these adhesion proteins, as well as advances in ways to identify and quantify the proteins, using proteomics, and then to analyse the data computationally, using bioinformatics.
The computational integration of multiple sets of adhesion proteins has now enabled the definition of a ‘meta-adhesome’, incorporating data from different cell types. From this analysis, a core set of 60 frequently identified proteins was identified, and this was termed a ‘consensus’ adhesome.
The assembly and disassembly of adhesion complexes is tightly regulated, to enable cells to attach to and detach from their surroundings, to migrate effectively and to control other cellular processes. But precisely how this happens is poorly understood.
Recent fluorescence microscopy analyses have led to a model of adhesion complex assembly in which adhesion proteins join adhesion complexes in a specific order. There appear to be roles for certain adhesion proteins at specific stages of adhesion complex formation. Quantification by proteomics of adhesion proteins during assembly and disassembly also revealed distinct patterns of protein recruitment, supporting and adding details to studies using microscopy.
This proposed system of adhesion complex assembly and disassembly might allow the rapid fine-tuning of adhesion signalling in cells.
But, despite their nametag, adhesion proteins are not just found associated with cell-matrix contacts. For example, the adhesion protein focal adhesion kinase (FAK) can be found in the nucleus of normal cells when they have undergone cellular stress.
FAK is also found in the nucleus of skin cancer cells, where it associates with packed DNA called chromatin.
It turns out that, in the nucleus, FAK profoundly influences the expression of genes. Using an integrative proteomic, bioinformatic and network approach, it was discovered that FAK binds to proteins in the nucleus that control gene expression. These proteins regulate the expression of chemokine genes, for example, which encode signalling proteins that are released by cells into their surroundings.
The release of chemokines, under the control of FAK, attracts regulatory T cells from the immune system. In skin cancer, this results in the cancer cells evading destruction by the immune system, which leads to tumour growth.
Importantly, there is no detectable nuclear FAK in normal skin cells called keratinocytes, suggesting that nuclear accumulation of FAK is linked to the pathology of skin cancer. This raises the possibility that nuclear FAK could be targeted therapeutically to treat cancer.
From a cell biological viewpoint, these and other data indicate that long-range signalling by adhesion proteins – at sites far from cell-matrix contacts – contribute to diverse cellular processes.
Understanding the precise relationships between multiple adhesion proteins, and how they work together to control cell behaviour, are important future priorities for research.
Only by understanding the networks of adhesion proteins and their interactions will we find out how cell adhesion signalling co-ordinates so many aspects of cell biology.
Citation Byron & Frame. Adhesion protein networks reveal functions proximal and distal to cell-matrix contacts. Curr. Opin. Cell Biol. 39, 93–100 (2016) HTML | PDF
This "News item" has been prepared by Dr Adam Byron and is reproduced here with author's permission. The original version is available on author's blog site: http://adambyron.com/2016/02/29/adhesion-protein-networks-home-and-away/

