Robin Allshire
Epigenetic mechanisms mediating antifungal resistance

Robin Allshire is a Wellcome Principal Research Fellow and Professor of Chromosome Biology at the University of Edinburgh. After graduating from Trinity College Dublin, Ireland in 1981 with a BA in Genetics, he obtained a Royal Commission for the Exhibition of 1851 scholarship and studied for a PhD at the MRC Mammalian Genome Unit, Edinburgh (MRC HGU) with Dr. Chris Bostock and Ed Southern. Following post-doctoral research at the MRC HGU with Prof. N.D. Hastie, he began his independent research career in 1989 at Cold Spring Harbor Laboratories, New York. He returned to a tenured position at MRC HGU from 1990 to 2002 during which he took a three month research sabbatical with Prof. Mitsuhiro Yanagida at Kyoto University.
In 2002, he moved to the Wellcome Centre for Cell Biology where he runs a dynamic research group as a Wellcome Principal Research Fellow (2002-2022). He was elected a member of the European Molecular Biology Organisation (EMBO) in 1998, a fellow of the Royal Society of Edinburgh (FRSE) in 2005 and a fellow of the Royal Society (FRS), London in 2011. He was awarded the Genetics Society (UK) Medal in 2013.
Lab members
Roberta Carloni, Andreas Fellas, Elisabeth Gaberdiel, Nitobe London, Alison Pidoux, Severina Pociunaite, Paresh Rana, Manu Shukla, Pin Tong, Ane Valera, Sharon White and Rebecca Yeboah
A simple overview of research in the Allshire Lab - Research in a Nutshell Videos
Epigenetic mechanisms mediating antifungal resistance
Antifungal resistance is increasing in prevalence, raising fungal-borne disease frequencies in humans and crops important for human well-being. The survival of fungi in harsh environments involves stress-sensing pathways that reprogram their proteomes. New environmental conditions, including global heating, can push opportunistic fungi to colonise novel niches, thus increasing their potential to become harmful pathogens. Effective antifungal treatments are limited in number precisely because fungi are adept at resisting challenges.
Resistance to fungicides/antifungal compounds can result from genetic mutations, however, it was unknown if resistance might also arise from heritable epigenetic changes mediated by post-translational modifications carried on histones in chromatin. Using the model fission yeast (Schizosaccharomyces pombe) fungal system, we discovered that heterochromatin island-mediated ‘epimutations’ confer resistance following exposure to external insults (Torres-Garcia et al. 2020; Figure A). Heterochromatin islands are formed by addition of methyl groups to lysine 9 of histone H3 (H3K9me) over regions of chromatin, resulting in reduced expression of underlying genes (Figure B). For example, epimutationmediated repression of the cup1+ gene encoding a mitochondrial LYR protein confers resistance through mitochondrial dysfunction (Figure C, D).
Transient ectopic H3K9me-dependent heterochromatin is normally rapidly erased by the counteracting H3K9 JmjC-domain Epe1 demethylase. Surprisingly, external insults such as antifungal compounds (e.g. caffeine, fluconazole) induce cleavage of Epe1 allowing heterochromatin islands to persist and confer resistance in selected lineages (Figure E). Unlike genetic mutations, such epimutations are unstable - causative heterochromatin islands, associated gene repression and resistance are lost in the absence of antifungal selection. Thus, epigenetic processes promote phenotypic plasticity so that wild-type cells adapt to unfavourable environments without irreversible genetic alterations.
We are exploiting fission yeast to define the mechanisms of epigenetic regulation that govern adaptation to challenging environments. The resulting findings will drive our investigations of processes governing the frequent emergence of antifungal resistance in divergent human (Cryptococcus neoformans) and plant (wheat; Zymoseptoria tritici) pathogens to identify and understand similarities and differences in the underlying processes.
Key questions
1. How are heterochromatin-dependent epimutations formed and maintained?
2. What features allow specific loci and individual cells to acquire epimutations and survive insults?
3. Do related epigenetic mechanisms mediate antifungal resistance in divergent pathogenic fungi?
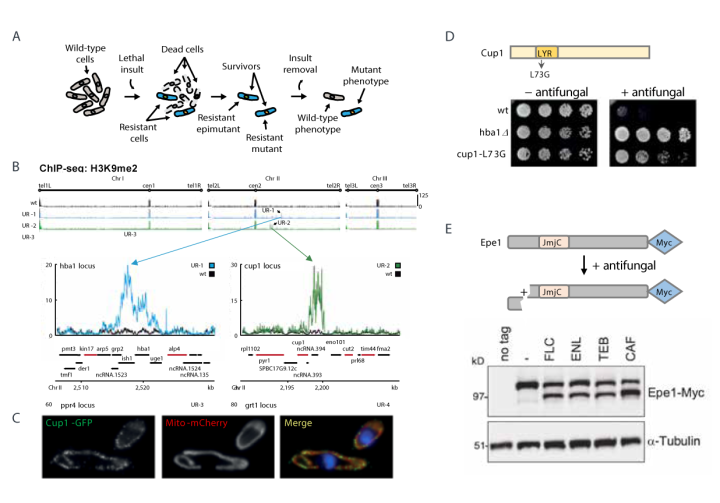
A. Model: Resistant isolates arise in fission yeast after insult exposure. Resistance can be mediated by changes in DNA (resistant mutants) or reversible, heterochromatin-based epimutations (resistant epimutants). Upon withdrawal of insult, epimutants lose heterochromatin islands, gene repression and resistance, reverting to wild-type (sensitive phenotype). In contrast, genetic mutants continue to exhibit the mutant resistant phenotype.
B. Unstable resistant epimutants UR-1 and UR-2 exhibit novel H3K9me-dependent heterochromatin islands compared to wildtype cells (wt). Repression of hba1+ and cup1+ genes confer caffeine or antifungal resistance in UR-1 and UR-2, respectively.
C. GFP-tagged Cup1 protein (cup1+ gene, UR-2) localises to mitochondria.
D. Mutation of a conserved leucine residue (L73G) in the Cup1 LYR domain confers antifungal resistance.
E. Exposure of fission yeast to clinical (FLC, Fluconazole) or agricultural (TEB, Tebuconazole; ENL, Enilconazole) antifungals, or caffeine (CAF) results in cleavage of Epe1 promoting heterochromatin island and resistant epimutation formation (Yaseen, White et al, BioRxiv doi.org/10.1101/2021.12.20.473483 ).
Selected publications
Fitz-James, M.H., Tong, P., Pidoux, A.L., Ozadam, H., Yang, L., White, S.A., Dekker, J., Allshire, R.C. (2020). Large domains of heterochromatin direct the formation of short mitotic chromosome loops. Elife 9, e57212. doi: 10.7554/eLife.57212.
Torres-Garcia, S., Yaseen, I., Shukla, M., Audergon, P.N.C.B., White, S.A., Pidoux, A.L., Allshire, R.C. (2020). Epigenetic gene silencing by heterochromatin primes fungal resistance. Nature 585, 453-458. doi: 10.1038/s41586-020-2706-x.
Staneva, D.P., Carloni, R., Auchynnikava, T., Tong, P., Rappsilber, J., Jeyaprakash A.A., Matthews K.R., Allshire, R.C. (2021) A systematic analysis of Trypanosoma brucei chromatin factors identifies novel protein interaction networks associated with sites of transcription initiation and termination. Genome Research 31:2138. doi: 10.1101/gr.275368.121.

