Matthew Swaffer
Effects of cell size on gene expression, biosynthesis and cell function

Matthew will join the Centre in August 2023.
Matthew is a Wellcome Trust CDA Fellow and incoming Group Leader at the Wellcome Centre for Cell Biology. Drawing on various sub-disciplines in cell, molecular and systems biology, Matthew's group aims to understand how and why different cellular processes and pathways are re-wired as cells grow and change their size.
After studying Natural Sciences at the University of Cambridge, Matthew obtained his PhD in Sir Paul Nurse's lab at the Francis Crick Institute working on Cyclin-Cdk substrate phosphorylation dynamics. In 2017 he moved to Stanford as a postdoc with Jan Skothiem where he developed quantitative functional genomics approaches to address the long-standing mystery of how global transcriptional output is scaled with cell size. Matthew will be establishing his own group at the WCB in 2023.
How does cell size impact gene expression, biosynthesis and cell function?
The size of a cell is one of its most basic and fundamental properties. While the significance of cell size for cellular physiology has been appreciated for many decades, we still have a surprisingly poor understanding of the mechanisms that couple the inner molecular workings of the cell to its size.
Core cellular processes must be coordinated with cell size to sustain a cell’s ability to grow and function optimally. For instance, protein and RNA amounts, as well as organelle volumes, increase continuously during the cell cycle in proportion to cell size, even though the template DNA genome does not (Figure A). This global scaling of macromolecules with size is critical as it ensures that the concentrations of enzymes and reactants are kept constant so that reactions and core cellular processes are maintained at a constant rate as cells grow and vary in size. As such, a major and long-standing question in cell biology has been: how is the production of macromolecules regulated to ensure this coupling with cell size?
We will be employing molecular and systems approaches to study how global gene expression, RNA processing, proteome homeostasis and cell division are regulated as cells grow and increase in size. Addressing these questions requires us to build a more quantitative view of how the central dogma plays out in vivo and this is one major theme of our work. To do this we will take an interdisciplinary approach that combines functional genomics, proteomics, imaging, and computational biology and utilise both yeast and mammalian cell lines as model systems.
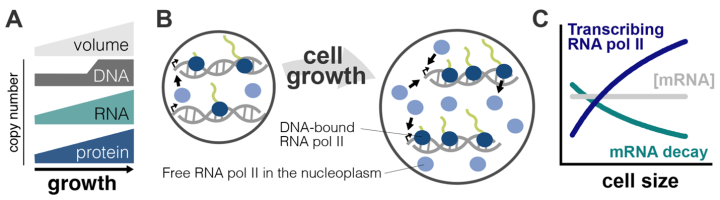
Recent and future research
We have recently shown that the genome-binding equilibrium kinetics of RNA polymerase II drives a global increase in mRNA transcription as cells grow and increase in size (Swaffer et al., 2022)(Figure B). However, this scaling is not directly proportional to size, which led us to discover that mRNA decay rates are also modulated to stabilise the transcriptome when cell size increases. This results in the proportional scaling of mRNA amounts with cell size and thereby ensures global mRNA concentration homeostasis (Figure C).
These results raise several important questions about how cellular biosynthesis is coordinated with cell size that we plan to pursue. What are the molecular mechanisms that adjust mRNA turnover in larger cells? What are the limits on this balance between transcription and decay and what are the consequences when such limits are reached? How do size-dependent changes in mRNA synthesis and turnover impact global proteome homeostasis? How does altered cell size impact other aspects of genome function including chromatin compaction and organisation? How conserved are these mechanisms across species and are similar processes at play to coordinate tRNA and rRNA production with the size-scaling of mRNA?
Selected publications
M. P. Swaffer, G. Marinov, H. Zheng, C. Tsui, A. W. Jones, J. Greenwood, A. Kundaje, W. Greenleaf, R. Reyes-Lamothe, J. M. Skotheim. RNA polymerase dynamics and mRNA stability feedback determine mRNA scaling with cell size. bioRixv 2021.09.20.461005 (2022)
M. C. Lanz, E. Zatulovskiy, M. P. Swaffer, L. Zhang, S. Zhang, D. S. You, G. K. Marinov, P. McAlpine, J. E. Elias, J. M. Skotheim. Increasing cell size remodels the proteome and promotes senescence. Molecular Cell, 82(17), 3255–3269.e8 (2022)
M. P. Swaffer, J. Kim, D. Chandler-Brown, M. Langhinrichs, G. Marinov, W. Greenleaf, A. Kundaje, K. M. Schmoller, J. M. Skotheim. Size-independent mRNA synthesis and chromatin-based partitioning mechanisms generate and maintain constant amounts of protein per cell. Molecular Cell, 81(23), 4861–4875.e7 (2021)
S. Xie, M. Swaffer, J. M. Skotheim. Eukaryotic Cell Size Control and Its Relation to Biosynthesis and Senescence. Annu Rev Cell Dev Biol. (2022)

