Juri Rappsilber
Cellular Tomography

Juri Rappsilber is a Wellcome Senior Research Fellow, Professor of Proteomics in the University of Edinburgh and Professor of Bioanalytics in the Technische Universität Berlin. His group develops tools to study the function, location, interactions and structure of proteins in cells. Most of the work involves cross-linking, mass spectrometry, machine learning and software development.
Juri Rappsilber took his PhD in Proteomics at the European Molecular Biology Laboratory (EMBL) in Heidelberg and Goethe Universität Frankfurt am Main in the lab of Matthias Mann and followed him as a postdoc to Odense, Denmark. In 2003 he relocated as principle investigator to the FIRC Institute of Molecular Oncology, Milan, which he left in 2006 to move to the University of Edinburgh. He became Wellcome Senior Research Fellow in 2009 and Professor of Proteomics in 2010. He has also held the post of Professor of Bioanalytics in Berlin since 2011.
Lab members
Colin Combe, Christos Spanos and Juan Zou
A simple explanation of research in the Rappsilber lab - Research in a Nutshell Videos
Cellular Tomography
Protein-protein interactions (PPIs) govern most cellular pathways and processes, and multiple technologies have emerged to systematically map them. Assessing the error of interaction networks has been a challenge hitherto, however. This has been true also for protein-protein interactions detected by crosslinking mass spectrometry. Crosslinking mass spectrometry is currently widening its scope from structural analyses of purified multi-protein complexes towards systemswide analyses of PPIs, to systematically reveal contact surfaces of the proteins. Using a carefully controlled large-scale analysis of Escherichia coli cell lysate, we demonstrated in 2021 that false-discovery rates (FDR) for PPIs identified by crosslinking mass spectrometry can be reliably estimated. The two key aspects for reliable error assessment in crosslinking are: (1) the separate handling of links that fall within proteins from those that fall between proteins, as the differ fundamentally in the size of the associated search spaces and consequently also in their random match behaviour (noise) and (2) assessing the error at the information level of interest (usually residue pairs or protein pairs, as opposed to the frequently used peptide-spectra matches). Applying these principles to our data using an open source tool that we made available, xiFDR, yielded an interaction network comprising 590 PPIs at 1% decoy-based PPI-FDR for E. coli. The structural information included in this network localises the binding site of the hitherto uncharacterised protein YacL to near the DNA exit tunnel on the RNA polymerase.
While this allowed to control the error in our data, we where left with limitations in the number of identified crosslinks. The incomplete and noisy information in the mass spectra of crosslinked peptides severely limits the numbers of protein– protein interactions that can be confidently identified. We therefore leveraged chromatographic retention time information to aid the identification of crosslinked peptides from mass spectra. Our Siamese machine learning model xiRT achieved highly accurate retention time predictions of crosslinked peptides in a multi-dimensional separation of crosslinked E. coli lysate. Importantly, supplementing the search engine score with retention time features led to a substantial increase in protein–protein interactions without affecting confidence. This approach is not limited to cell lysates and multidimensional separation but also improved considerably the analysis of crosslinked multiprotein complexes with a single chromatographic dimension, as we could demonstrate for an analysis of the Fanconi anemia complex (see figure). Retention times are a powerful complement to mass spectrometric information to increase the sensitivity of crosslinking mass spectrometry analyses.
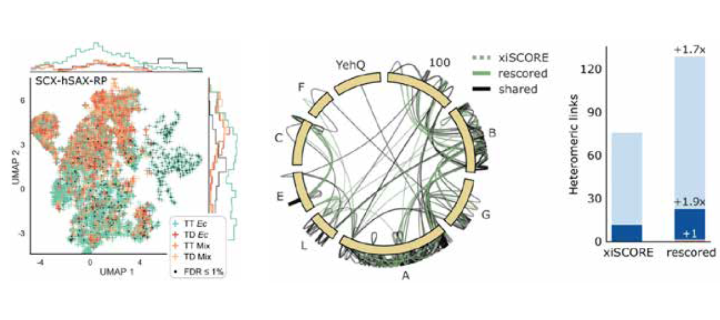
Retention time data complements substantially the currently exclusively used mass spectrometric evidence for the identification of crosslinks between proteins.
Left: The combined retention information of crosslinked peptides from three different chromatography modes suffices to effectively separate plausible identifications (green) from modelled noise (all other colours) in a crosslink analysis of E. coli lysate.
Middle: Crosslink network from the Fanconi anemia complex analysis, shown in the circular view. Unique residue pairs from xiSCORE (gray), after rescoring (green), and shared (black) between these analyses are depicted (1% residue-pair FDR). Proteins associated to the Fanconi anemia core complex are indicated with their gene name suffix. The E. coli protein YehQ represents a match from the entrapment database.
Right: Quantitative assessment of residue-pairs with and without rescoring, and including calculated distances in the model (all, light blue; ≤35Å, blue; >35Å red), showing a gain in 70% on identified crosslinks between proteins.
Selected publications
O’Reilly, F.J., Xue, L., Graziadei, A., Sinn, L., Lenz, S., Tegunov, D., Blötz, C., Singh, N., Hagen, W.J.H., Cramer, P., Stülke, J., Mahamid, J., Rappsilber, J. (2020). In-cell architecture of an actively transcribing-translating expressome. Science 369, 554–557.
Giese, S.H., Sinn, L.R., Wegner, F., and Rappsilber, J. (2021). Retention time prediction using neural networks increases identifications in crosslinking mass spectrometry. Nat. Commun. 12, 3237.
Lenz, S., Sinn, L.R., O’Reilly, F.J., Fischer, L., Wegner, F., and Rappsilber, J. (2021). Reliable identification of protein-protein interactions by crosslinking mass spectrometry. Nat. Commun. 12, 3564.

