Bill Earnshaw
The role of non-histone proteins in chromosome structure and function during mitosis

William Earnshaw moved to Edinburgh in 1996 as a Wellcome Trust Principal Research Fellow, which he remains today. He graduated summa cum laude from Colby College in Waterville Maine in 1972, then completed a Ph.D. with Jonathan King at MIT in 1977 after a brief stint in the US Air Force. Postdoctoral training in Cambridge with Aaron Klug, Tony Crowther and Ron Laskey and in Geneva with Ulrich Laemmli was followed by 13 years at the Johns Hopkins School of Medicine. Throughout his career, his studies have focused on the packaging and segregation of chromosomes during cell division. Achievements of his team during his time in Edinburgh include identification of the chromosomal passenger complex, construction of the first human synthetic artificial chromosome and the use of multidisciplinary approaches to study the organisation and formation of vertebrate mitotic chromosomes. He has been elected to EMBO, the Royal Society of Edinburgh, the Academy of Medical Sciences and the Royal Society of London. He co-authors the textbook Cell Biology with Tom Pollard, Graham Johnson and Jennifer Lippincott-Schwartz (4th edition published in 2023). For more information see the Earnshaw lab website.
Lab members
Fernanda Cisneros-Soberanis, Lorenza Di Pompeo, Moonmoon Deb, Natalia Kochanova, Bram Prevo, Nina Pučeková, Caitlin Reid, Lucy Remnant, Itaru Samejima and Kumiko Samejima
A simple explanation of research in the Earnshaw lab - Research in a Nutshell Videos
The role of non-histone proteins in chromosome structure and function during mitosis
Over the past year, much of our research focused on structural dynamics in chromatin during the transition of cells from G2 phase into mitosis, the role of SMC proteins in mitotic chromosome formation and structure and the structure and assembly of the chromosome periphery.
One highlight was the publication of a study that has been ongoing for several years in which Itaru examined the changes in protein association with chromatin during synchronous mitotic entry. This study used Chromatin Enrichment for Proteomics (ChEP), a method developed by Georg Kustatscher when he was a postdoc with Juri Rappsilber, and was a collaboration between the three labs. We discovered that the earliest events of prophase appear to primarily involve changes in RNA processing in nuclei as well as changes in interactions with the nuclear envelope and pores. All of these events begin before chromatin condensation is visible, and the study was only made possible by using the chemicalgenetic system for synchronous mitotic entry developed by Kumiko.
We are currently writing up the results of our long-running study of interactions between cohesin and condensin during mitotic chromosome formation. This is a truly interdisciplinary collaboration with the groups of Job Dekker, Leonid Mirny and Anton Goloborodko. We do the genetics, cell biology and imaging. They do Hi-C and polymer modelling, respectively. We have discovered that cohesin has a significant effect on mitotic chromosome structure that has been previously overlooked and gained surprising new insights into the organisation of the chromatin fiber in chromosomes. Kumiko has made many genomic knock-in cell lines, performed the cell synchrony and carried out extensive light microscopy analysis. Fernanda and Nina have been performing serial block face scanning electron microscopy with our collaborators Ian Prior and Alison Beckett in Liverpool. Itaru has been doing ChEP and Moonmoon has been doing ChIP - both to quantitate the amounts of condensin and cohesin on the chromosomes during mitotic entry.
In other ongoing work, Lucy and Fernanda are studying the enigmatic protein Ki-67 and the RNA/protein-rich mitotic chromosome periphery compartment (MCPC), Lorenza is performing a structure/function analysis on CENP-V, Caitlin is studying the role of topo IIβ in chromosome formation, Natalia is using proteomics to look at protein conformations and interactions during mitotic entry, and Bram is developing new ways to image chromosomes.
Our work is supported by a Wellcome Principal Research Fellowship and by the Centre for Mammalian Synthetic Biology.
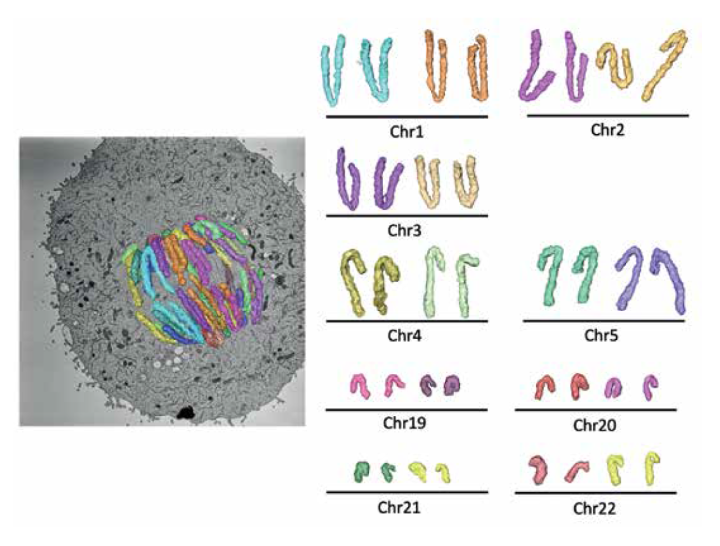
Three-dimensional reconstruction of an anaphase human RPE1 cell. Left, projection of the three-dimensional reconstruction superimposed on an orthoslice from the electron microscopy map. Corresponding sister chromatids have the same colours. Right, partial karyotype with individual sister chromatids (identified by size and centromere position) extracted from the map and displayed next to their sisters. Sample preparation and modeling in AMIRA by Fernanda Cisneros-Soberanis. Serial block face scanning electron microscopy by Alison Beckett and Ian Prior, University of Liverpool.
Selected publications
Samejima, I., C. Spanos, K. Samejima, J. Rappsilber, G. Kustatscher & W.C. Earnshaw. (2022). Mapping the invisible chromatin transactions of prophase chromosome remodelling. MOL. CELL 82:696-708; PMID: 35090599; PMCID: 8823707; DOI: 10.1016/j. molcel.2021.12.039.
Paulson JR, Hudson DF, Cisneros-Soberanis F, Earnshaw WC. (2021). Mitotic chromosomes. SEMIN CELL DEV BIOL. 117:7-29. PMID: 33836947; PMC8406421; DOI:10.1016/j.semcdb.2021.03.014. Pesenti,E., M. Liskovykh, K. Okazaki, A. Mallozzi, C. Reid, M.A. Abad, A.A. Jeyaprakash, N. Kouprina, V. Larionov, H. Masumoto, W.C.
Earnshaw. (2020) Analysis of Complex DNA Rearrangements During Early Stages of HAC Formation. ACS SYNTH BIOL. 9:3267-3287. PMID: 33289546; PMC7754191; DOI: 10.1021/acssynbio.0c00326.

