Breakthrough article from Greiss lab published in Nucleic Acids Research
Using a quadruplet codon to expand the genetic code of an animal
This work introduces a system for the use of quadruplet codons to direct incorporation of non-canonical amino acids in vivo in an animal, the nematode worm Caenorhabditis elegans.
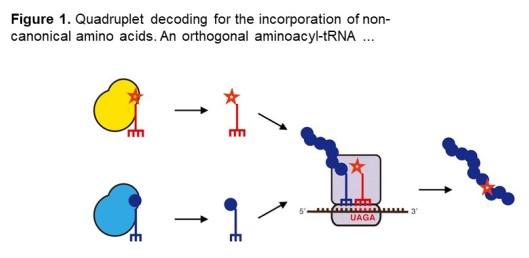
In the paper researchers construct 'hybrid' tRNAs to decode UAGA quadruplet codons and incorporate photocaged amino acids in C. elegans. They use photocaged lysine to express photocaged Cre recombinase for optical control of gene expression. Here the UAGA quadruplet works as efficiently as a UAG triplet. The quadruplet is then used to incorporate photocaged cysteine into a caspase for light induced ablation of C. elegans neurons.
Their approach will facilitate the routine adoption of quadruplet decoding for genetic code expansion in eukaryotic cells and multicellular organisms.
Congratulations to Zhiyan Xi and everyone involved: Lloyd Davis, Kieran Baxter, Ailish Tynan, Angeliki Goutou and Sebastian Greiss.
SynthSys member Sebastian Greiss is a Chancellor’s fellow with the Centre for Discovery Brain Sciences. The Greiss lab develops and uses tools based on unnatural amino acids to probe neuronal circuits in C. elegans worms.
Read more

